Integration of pygcc with EQ3/6
Import pygcc, EQ3/6 environment and other modules
All files used in this tutorial can be downloaded from https://bitbucket.org/Tutolo-RTG/pygcc/src/master/docs/
Download output_reader into your working directory
import os, subprocess, re, numpy as np, pandas as pd, sys
import shutil
from pygcc.pygcc_utils import *
import pygcc
print(pygcc.__version__)
from output_reader import read_eq36output
1.2.0
Set current working directory and EQ3/6 working directory. This is because EQ3/6 can only run from its working directory
eq_dir = "C:\Eq3_6v8.0a\work_db"
if os.path.exists(eq_dir) == False:
os.makedirs(os.path.join(eq_dir))
current_dir = os.getcwd()
output_folder = 'Vent_spec'
if os.path.exists(os.path.join(os.getcwd(), output_folder)) == False:
os.makedirs(os.path.join(os.getcwd(), output_folder))
# function to copy file between source and destination directory
def copyfile(source_dir, destination_dir):
# Source path
source = source_dir
# Destination path
destination = destination_dir
try:
shutil.copy(source, destination)
print("File copied successfully.")
# If source and destination are same
except shutil.SameFileError:
print("Same files are in the Source and destination path.")
# If there is any permission issue
except PermissionError:
print("Permission denied.")
# For other errors
except:
print("Error occurred while copying file.")
Load filtered vent fluid data
filename = './MARHYS_DB_2_0_filter.csv'
df = pd.read_csv(filename)
i = 2
df.loc[i, :]
Sample-ID EPR 21°N-OBS-1985
Geol_setting Mid-oceanic spreading center
Rock_type_primary Basalt
Depth 2500
Temp 340
pH 3.4
XH2S 7.6
Cl 500.0
Na 439.0
Fe 1530
Mn 1024
H2 0.01
K 23.5
Ca 15.6
Si 17.6
Cu 0.0
Press 251.945325
Name: 2, dtype: object
Here we will work with EPR 21 N sample to run speciation and warm up the fluid from 25 °C to 340 °C
Generate the database for the vent fluid with pygcc and run the database through EQ3/6 preprocessing
We initialize the inputs and generate the database
P = np.round(df.loc[i,:].Press)
Temp = df.loc[i,:].Temp; npH = df.loc[i,:].pH
# multiplication was done to convert the concentrations from mmol/kg and umol/kg to mol/kg
nCl = df.loc[i,:].Cl*1e-3; nNa = df.loc[i,:].Na*1e-3
nH2S = df.loc[i,:].XH2S*1e-3; nFe = df.loc[i,:].Fe*1e-6
nMn = df.loc[i,:].Mn*1e-6; nH2 = df.loc[i,:].H2*1e-6
nK = df.loc[i,:].K*1e-3; nCu = df.loc[i,:].Cu*1e-6;
nCa = df.loc[i,:].Ca*1e-3; nSi = df.loc[i,:].Si*1e-3
write_database(T = [20, Temp], P = P, solid_solution = True, objdb = './data0',
clay_thermo = 'Yes', dataset = 'eq36', sourcedb = './database/data0.geo') #
Success, your new EQ3/6 database is ready for download
<pygcc.pygcc_utils.write_database at 0x7fc783f2efd0>
Copy the generated database to the EQ3/6 work environment specified above and run through eqpt for preprocessing
copyfile(current_dir + '/output/EQ36/data0.geo', eq_dir + '/data0.pym')
File copied successfully.
# change the working directory to EQ3/6 work environment
os.chdir(eq_dir)
# load EQ3/6 environment
os.environ["EQ36CO"] = "C:\Eq3_6v8.0a\Bin"
os.environ["EQ36DA"] = eq_dir
# check the "eqptout.txt" for the output of the database preprocessing
f = open(current_dir + "/%s/eqptout.txt" % output_folder, "w")
subprocess.run([r'C:/Eq3_6v8.0a/bin/runeqpt', 'pym'], stdout = f)
f.close()
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_268/326673575.py in <module>
1 # check the "eqptout.txt" for the output of the database preprocessing
2 f = open(current_dir + "/%s/eqptout.txt" % output_folder, "w")
----> 3 subprocess.run([r'C:/Eq3_6v8.0a/bin/runeqpt', 'pym'], stdout = f)
4 f.close()
~/.pyenv/versions/3.7.9/lib/python3.7/subprocess.py in run(input, capture_output, timeout, check, *popenargs, **kwargs)
486 kwargs['stderr'] = PIPE
487
--> 488 with Popen(*popenargs, **kwargs) as process:
489 try:
490 stdout, stderr = process.communicate(input, timeout=timeout)
~/.pyenv/versions/3.7.9/lib/python3.7/subprocess.py in __init__(self, args, bufsize, executable, stdin, stdout, stderr, preexec_fn, close_fds, shell, cwd, env, universal_newlines, startupinfo, creationflags, restore_signals, start_new_session, pass_fds, encoding, errors, text)
798 c2pread, c2pwrite,
799 errread, errwrite,
--> 800 restore_signals, start_new_session)
801 except:
802 # Cleanup if the child failed starting.
~/.pyenv/versions/3.7.9/lib/python3.7/subprocess.py in _execute_child(self, args, executable, preexec_fn, close_fds, pass_fds, cwd, env, startupinfo, creationflags, shell, p2cread, p2cwrite, c2pread, c2pwrite, errread, errwrite, restore_signals, start_new_session)
1549 if errno_num == errno.ENOENT:
1550 err_msg += ': ' + repr(err_filename)
-> 1551 raise child_exception_type(errno_num, err_msg, err_filename)
1552 raise child_exception_type(err_msg)
1553
FileNotFoundError: [Errno 2] No such file or directory: 'C:/Eq3_6v8.0a/bin/runeqpt': 'C:/Eq3_6v8.0a/bin/runeqpt'
Replace species and concentration in an existing eq3 file
# change the working directory back to current work environment to do some calculations and build input files
os.chdir(current_dir)
# Switch basis between HS- and SO4--
lst = ['|Special Basis Switches (for model definition only) | (nsbswt) |\n',
'|------------------------------------------------------------------------------|\n',
'|Replace |SO4-- | (usbsw(1,n)) |\n',
'| with |HS- | (usbsw(2,n)) |\n',
'|------------------------------------------------------------------------------|\n']
lst
['|Special Basis Switches (for model definition only) | (nsbswt) |\n',
'|------------------------------------------------------------------------------|\n',
'|Replace |SO4-- | (usbsw(1,n)) |\n',
'| with |HS- | (usbsw(2,n)) |\n',
'|------------------------------------------------------------------------------|\n']
# Set H2(aq) as the redox constraint
lst2 = ['|Default redox constraint (irdxc3): |\n',
'| [ ] (-3) Use O2(g) line in the aqueous basis species block |\n',
'| [ ] (-2) pe (pe units) | 0.00000E+00| (pei) |\n',
'| [ ] (-1) Eh (volts) | 5.00000E-01| (ehi) |\n',
'| [ ] ( 0) Log fO2 (log bars) |-7.00000E-01| (fo2lgi) |\n',
'| [x] ( 1) Couple (aux. sp.) |H2(aq) | (uredox) |\n',
'|------------------------------------------------------------------------------|\n']
lst2
['|Default redox constraint (irdxc3): |\n',
'| [ ] (-3) Use O2(g) line in the aqueous basis species block |\n',
'| [ ] (-2) pe (pe units) | 0.00000E+00| (pei) |\n',
'| [ ] (-1) Eh (volts) | 5.00000E-01| (ehi) |\n',
'| [ ] ( 0) Log fO2 (log bars) |-7.00000E-01| (fo2lgi) |\n',
'| [x] ( 1) Couple (aux. sp.) |H2(aq) | (uredox) |\n',
'|------------------------------------------------------------------------------|\n']
ion_lst = ['Fe++', 'Na+', 'H+', 'Cl-', 'HS-', 'Mn++', 'K+', 'Ca++', 'SiO2(aq)', 'Cu++', 'H2(aq)'] # , 'H2S(aq)'
conc_lst = [nFe, nNa, npH, nCl, nH2S, nMn, nK, nCa, nSi, nCu, nH2]
lst3 = ['|Aqueous Basis Species/Constraint Species |Conc., etc. |Units/Constraint|\n',
'| (uspeci(n)/ucospi(n)) | (covali(n))|(ujf3(jflgi(n)))|\n',
'|------------------------------------------------------------------------------|\n']
for i, j in zip(ion_lst, conc_lst):
units = 'Molality ' if i != 'H+' else 'pH '
lst3.append('|%-10s | %.5e|%s|\n' % (i, j, units))
lst3.append('|------------------------------------------------------------------------------|\n')
lst3
['|Aqueous Basis Species/Constraint Species |Conc., etc. |Units/Constraint|\n',
'| (uspeci(n)/ucospi(n)) | (covali(n))|(ujf3(jflgi(n)))|\n',
'|------------------------------------------------------------------------------|\n',
'|Fe++ | 1.53000e-03|Molality |\n',
'|Na+ | 4.39000e-01|Molality |\n',
'|H+ | 3.40000e+00|pH |\n',
'|Cl- | 5.00000e-01|Molality |\n',
'|HS- | 7.60000e-03|Molality |\n',
'|Mn++ | 1.02400e-03|Molality |\n',
'|K+ | 2.35000e-02|Molality |\n',
'|Ca++ | 1.56000e-02|Molality |\n',
'|SiO2(aq) | 1.76000e-02|Molality |\n',
'|Cu++ | 1.00000e-13|Molality |\n',
'|H2(aq) | 1.00000e-08|Molality |\n',
'|------------------------------------------------------------------------------|\n']
# open an existing sample file and a new in put file
fin = open("swmaj_ex.3i", "r")
fout = open("./%s/swmaj.3i" % output_folder, "w")
# read and paste line by line until the beginning of the "Special Basis Switches" block
for i in range(1000):#
s = fin.readline()
if s.strip('|').startswith("Special Basis Switches"):
break
fout.writelines(s)
# paste the new "Special Basis Switches" block
fout.writelines(lst)
# read and skip pasting until the end of the "Special Basis Switches" block
for i in range(1000):#
s = fin.readline()
if s.strip('|').startswith("Temperature (C)"):
break
fout.writelines(s)
# read and paste line by line until the beginning of the "Default redox constraint" block
for i in range(1000):#
s = fin.readline()
if s.strip('|').startswith("Default redox constraint"):
break
fout.writelines(s)
# paste the new "Special Basis Switches" block
fout.writelines(lst2)
# read and skip pasting until the end of the "Default redox constraint" block and
# the beginning of the "Aqueous Basis Species" block
for i in range(1000):#
s = fin.readline()
if s.strip('|').startswith("Aqueous Basis Species/Constraint Species"):
break
# paste the new "Aqueous Basis Species" block
fout.writelines(lst3)
# read and skip pasting until the end of the "Aqueous Basis Species" block
for i in range(1000):#
s = fin.readline()
if s.strip('* ').startswith("Valid jflag strings"):
break
fout.writelines(s)
# read and paste the rest of the files
for i in range(1000):#
s = fin.readline()
fout.writelines(s)
fout.close()
fin.close()
Copy the new eq3 input file to the EQ3/6 work environment and run eq3
copyfile(current_dir + '/%s/swmaj.3i' % output_folder, eq_dir + '/swmaj.3i')
File copied successfully.
# change the working directory to EQ3/6 work environment
os.chdir(eq_dir)
# check the "eq3out.txt" for the eq3 run details
f = open(current_dir + "/%s/eq3out.txt" % output_folder, "w")
subprocess.run([r'C:\Eq3_6v8.0a/bin/runeq3', 'pym', 'swmaj.3i'], stdout = f)
f.close()
Generate a new eq6 file to run speciation and warm up temperature
# change the working directory back to current work environment to build input files
os.chdir(current_dir)
Descript = 'Heat ventfluid from 25C to %sC' % Temp
# open an existing sample file, a new input file and the copied eq3 output file
fin = open("swmaj_ex.6i", "r")
fout = open("./%s/swmaj.6i" % output_folder, "w")
f3p = open(eq_dir + "/swmaj.3p", "r")
# read and paste line by line until the beginning of the first "Linear tracking in Xi" in the "Temperature option" block
for i in range(1000):#
s = fin.readline()
if s.strip('| ').startswith("[x] ( 1) Linear tracking in Xi"):
break
if s.strip('|').startswith('EQ6 input file name'):
line = '%s= %-11s |\n' % (s.split('=')[0],
'swmaj.6i')
fout.writelines(line)
elif s.strip('|').startswith('Description'):
line = '%s= %-45s |\n' % (s.split('=')[0], Descript)
fout.writelines(line)
else:
fout.writelines(s)
#create the new "Linear tracking in Xi" subblock and paste the new subblock
derivative = (Temp - 25)/1
lst = ['| [x] ( 1) Linear tracking in Xi: |\n',
'| Base Value (C) | 2.50000E+01| (tempcb) |\n',
'| Derivative | %.5e| (ttk(1)) |\n' % derivative]
fout.writelines(lst)
# read and skip pasting until the end of the "Linear tracking in Xi" subblock
for i in range(1000):#
s = fin.readline()
if s.strip('|').lstrip().startswith("Derivative"):
break
# read and paste line by line until the beginning of the "eq3 output" input
for i in range(10000):#
s = fin.readline()
if s.strip('* ').startswith("Start of the bottom half of the INPUT file"):
break
fout.writelines(s)
# paste the new eq3 output
for i in range(1000):#
s = f3p.readline()
fout.writelines(s)
fout.close()
fin.close()
f3p.close()
Copy the new eq6 input file to the EQ3/6 work environment and run eq6
copyfile(current_dir + '/%s/swmaj.6i' % output_folder, eq_dir + '/swmaj.6i')
File copied successfully.
# change the working directory to EQ3/6 work environment
os.chdir(eq_dir)
# check the "eq6out.txt" for the eq6 run details
f = open(current_dir + "/%s/eq6out.txt" % output_folder, "w")
subprocess.run([r'C:\Eq3_6v8.0a/bin/runeq6', 'pym', 'swmaj.6i'], stdout = f)
f.close()
Results processing
data = {}
rerun = []
path = eq_dir + '/'
files = os.listdir(path)
for i, f in enumerate(files):
if f.endswith('.6o'):
size = os.path.getsize(path + f)/(1024) #size in KB
if size > 2:
data[f[:-3]] = read_eq36output(path + f)
else:
rerun.append(f)
print(rerun)
[]
data_df = data['swmaj']
# data_df
data_df.columns
Index(['Xi', 'pH', 'Temperature_C', 'Pressure_bars', 'Water_mass_g',
'Rock_mass_g', 'M_Ca++', 'M_CaCl+', 'M_CaCl2(aq)', 'M_CaHSiO3+',
'M_CaOH+', 'M_CaSO4(aq)', 'M_Cl-', 'M_Cu+', 'M_CuCl(aq)', 'M_CuCl2-',
'M_CuCl3--', 'M_Fe++', 'M_Fe+++', 'M_FeCl+', 'M_FeCl++', 'M_FeCl2(aq)',
'M_FeO(aq)', 'M_FeO+', 'M_FeO2-', 'M_FeOH+', 'M_FeOH++', 'M_H+',
'M_H2(aq)', 'M_H2S(aq)', 'M_H2S2O3(aq)', 'M_HFeO2(aq)', 'M_HFeO2-',
'M_HS-', 'M_HS2O3-', 'M_HSO3-', 'M_HSO4-', 'M_HSiO3-', 'M_K+',
'M_KCl(aq)', 'M_KHSO4(aq)', 'M_KOH(aq)', 'M_KSO4-', 'M_Mn++', 'M_MnCl+',
'M_MnO(aq)', 'M_MnOH+', 'M_MnSO4(aq)', 'M_Na+', 'M_NaCl(aq)',
'M_NaHSiO3(aq)', 'M_NaOH(aq)', 'M_NaSO4-', 'M_OH-', 'M_S2--',
'M_S2O3--', 'M_S3--', 'M_S4--', 'M_S5--', 'M_SO2(aq)', 'M_SO4--',
'M_SiO2(aq)', 'TotM_Ca++', 'TotM_Cl-', 'TotM_Cu++', 'TotM_Fe++',
'TotM_H+', 'TotM_H2O', 'TotM_HS-', 'TotM_K+', 'TotM_Mn++', 'TotM_Na+',
'TotM_O2(g)', 'TotM_SiO2(aq)', 'SI_Alabandite', 'SI_Anhydrite',
'SI_Bornite', 'SI_Chalcedony', 'SI_Chalcocite', 'SI_Chalcopyrite',
'SI_Coesite', 'SI_Covellite', 'SI_Cristobalite(beta)', 'SI_Fayalite',
'SI_FeO', 'SI_Ferrosilite', 'SI_Halite', 'SI_Hematite', 'SI_Magnetite',
'SI_Minnesotaite', 'SI_Pyrite', 'SI_Pyrrhotite', 'SI_Quartz',
'SI_SiO2(am)', 'SI_Sylvite'],
dtype='object')
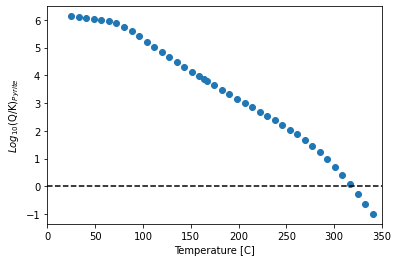
Plot pyrite saturation with Temperature
import matplotlib.pyplot as plt
y = data_df.SI_Pyrite
x = data_df.Temperature_C
plt.figure()
plt.scatter(x, y)
plt.plot([0, 350], [0, 0], 'k--')
plt.xlim([0, 350])
plt.xlabel('Temperature [C]')
plt.ylabel('$Log_{10}$(Q/K)$_{Pyrite}$')
Text(0, 0.5, '$Log_{10}$(Q/K)$_{Pyrite}$')
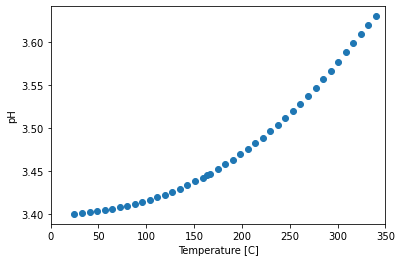
plot pH with Temperature
y = data_df.pH
x = data_df.Temperature_C
plt.figure()
sc = plt.scatter(x, y)
plt.xlim([0, 350])
plt.xlabel('Temperature [C]')
plt.ylabel('pH')
Text(0, 0.5, 'pH')